


Next: The Offset Structure
Up: Accessing a Darwin Sequence
Previous: Accessing a Darwin Sequence
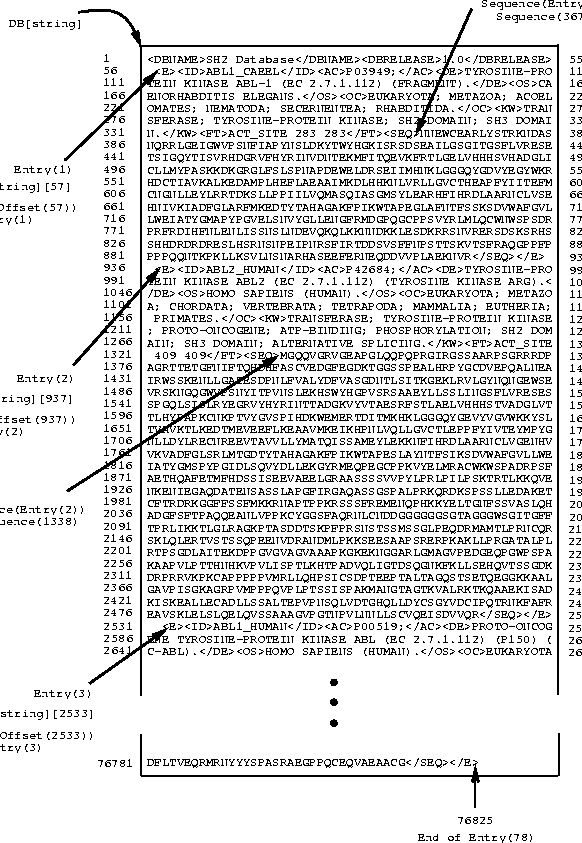
The entire database Sample/SH2 is stored in Darwin as a string of length DB[TotEntries]. Figure ![[*]](cross_ref_motif.gif) gives a graphical view of how information is organized internally.
DB[string] points to the beginning of this name.
Recall that each entry from a sequence database in Darwin is wrapped
in the SGML tags <E>, </E>. To extract the entire
contents of an entry, we use the Entry structured type.
gives a graphical view of how information is organized internally.
DB[string] points to the beginning of this name.
Recall that each entry from a sequence database in Darwin is wrapped
in the SGML tags <E>, </E>. To extract the entire
contents of an entry, we use the Entry structured type.
> ReadDb('Sample/SH2');
> first := Entry(1);
first := Entry(1)
> second := Entry(2);
second := Entry(2)
> last_three := Entry(76, 77, 78);
last_three := Entry(76,77,78)
> print(first);
<E><ID>ABL1_CAEEL</ID><AC>P03949;</AC><DE>TYROSINE-PROTEIN
KINASE ABL-1 (EC 2.7.1.112) (FRAGMENT).</DE><OS>CAENORHABD
ITIS ELEGANS.</OS><OC>EUKARYOTA; METAZOA; ACOELOMATES; NEM
ATODA; SECERNENTEA; RHABDITIDA.</OC><KW>TRANSFERASE; TYROS
INE-PROTEIN KINASE; SH2 DOMAIN; SH3 DOMAIN.</KW><FT>ACT_SI
TE 283 283</FT><SEQ>NNEWCEARLYSTRKNDASNQRRLGEIGWVPSNFIAPYN
SLDKYTWYHGKISRSDSEAILGSGITGSFLVRESETSIGQYTISVRHDGRVFHYRINV
DNTEKMFITQEVKFRTLGELVHHHSVHADGLICLLMYPASKKDKGRGLFSLSPNAPDE
WELDRSEIIMHNKLGGGQYGDVYEGYWKRHDCTIAVKALKEDAMPLHEFLAEAAIMKD
LHHKNLVRLLGVCTHEAPFYIITEFMCNGNLLEYLRRTDKSLLPPIILVQMASQIASG
MSYLEARHFIHRDLAARNCLVSEHNIVKIADFGLARFMKEDTYTAHAGAKFPIKWTAP
EGLAFNTFSSKSDVWAFGVLLWEIATYGMAPYPGVELSNVYGLLENGFRMDGPQGCPP
SVYRLMLQCWNWSPSDRPRFRDIHFNLENLISSNSLNDEVQKQLKKNNDKKLESDKRR
SNVRERSDSKSRHSSHHDRDRDRESLHSRNSNPEIPNRSFIRTDDSVSFFNPSTTSKV
TSFRAQGPPFPPPPQQNTKPKLLKSVLNSNARHASEEFERNEQDDVVPLAEKNVR</S
EQ></E>
> print(second);
<E><ID>ABL2_HUMAN</ID><AC>P42684;</AC><DE>TYROSINE-PROTEIN
KINASE ABL2 (EC 2.7.1.112) (TYROSINE KINASE ARG).</DE><OS
>HOMO SAPIENS (HUMAN).</OS><OC>EUKARYOTA; METAZOA; CHORDAT
A; VERTEBRATA; TETRAPODA; MAMMALIA; EUTHERIA; PRIMATES.</O
C><KW>TRANSFERASE; TYROSINE-PROTEIN KINASE; PROTO-ONCOGENE
; ATP-BINDING; PHOSPHORYLATION; SH2 DOMAIN; SH3 DOMAIN; AL
TERNATIVE SPLICING.</KW><FT>ACT_SITE 409 409</FT><SEQ>MGQQ
VGRVGEAPGLQQPQPRGIRGSSAARPSGRRRDPAGRTTETGFNIFTQHDHFASCVEDG
FEGDKTGGSSPEALHRPYGCDVEPQALNEAIRWSSKENLLGATESDPNLFVALYDFVA
SGDNTLSITKGEKLRVLGYNQNGEWSEVRSKNGQGWVPSNYITPVNSLEKHSWYHGPV
SRSAAEYLLSSLINGSFLVRESESSPGQLSISLRYEGRVYHYRINTTADGKVYVTAES
RFSTLAELVHHHSTVADGLVTTLHYPAPKCNKPTVYGVSPIHDKWEMERTDITMKHKL
GGGQYGEVYVGVWKKYSLTVAVKTLKEDTMEVEEFLKEAAVMKEIKHPNLVQLLGVCT
LEPPFYIVTEYMPYGNLLDYLRECNREEVTAVVLLYMATQISSAMEYLEKKNFIHRDL
AARNCLVGENHVVKVADFGLSRLMTGDTYTAHAGAKFPIKWTAPESLAYNTFSIKSDV
WAFGVLLWEIATYGMSPYPGIDLSQVYDLLEKGYRMEQPEGCPPKVYELMRACWKWSP
ADRPSFAETHQAFETMFHDSSISEEVAEELGRAASSSSVVPYLPRLPILPSKTRTLKK
QVENKENIEGAQDATENSASSLAPGFIRGAQASSGSPALPRKQRDKSPSSLLEDAKET
CFTRDRKGGFFSSFMKKRNAPTPPKRSSSFREMENQPHKKYELTGNFSSVASLQHADG
FSFTPAQQEANLVPPKCYGGSFAQRNLCNDDGGGGGGSGTAGGGWSGITGFFTPRLIK
KTLGLRAGKPTASDDTSKPFPRSNSTSSMSSGLPEQDRMAMTLPRNCQRSKLQLERTV
STSSQPEENVDRANDMLPKKSEESAAPSRERPKAKLLPRGATALPLRTPSGDLAITEK
DPPGVGVAGVAAAPKGKEKNGGARLGMAGVPEDGEQPGWPSPAKAAPVLPTTHNHKVP
VLISPTLKHTPADVQLIGTDSQGNKFKLLSEHQVTSSGDKDRPRRVKPKCAPPPPPVM
RLLQHPSICSDPTEEPTALTAGQSTSETQEGGKKAALGAVPISGKAGRPVMPPPQVPL
PTSSISPAKMANGTAGTKVALRKTKQAAEKISADKISKEALLECADLLSSALTEPVPN
SQLVDTGHQLLDYCSGYVDCIPQTRNKFAFREAVSKLELSLQELQVSSAAAGVPGTNP
VLNNLLSCVQEISDVVQR</SEQ></E>
> print(last_three);
E><ID>YRK_CHICK</ID><AC>Q02977;</AC><DE>PROTO-ONCOGENE TYR
OSINE-PROTEIN KINASE YRK (EC 2.7.1.112) (P60-YRK) (YES REL
ATED KINASE).</DE><OS>GALLUS GALLUS (CHICKEN).</OS><OC>EUK
...
E><ID>ZA70_HUMAN</ID><AC>P43403;</AC><DE>TYROSINE-PROTEIN
KINASE ZAP-70 (EC 2.7.1.112) (70 KD ZETA-ASSOCIATED PROTEI
N).</DE><OS>HOMO SAPIENS (HUMAN).</OS><OC>EUKARYOTA; METAZ
...
<E><ID>ZA70_MOUSE</ID><AC>P43404;</AC><DE>TYROSINE-PROTEIN
KINASE ZAP-70 (EC 2.7.1.112) (70 KD ZETA-ASSOCIATED PROTE
IN).</DE><OS>MUS MUSCULUS (MOUSE).</OS><OC>EUKARYOTA; META
...
We can isolate the contents of a specific SGML tag by including the
tag in single quotes and square brackets.
> first['ID']; # get the identification tag of the 1st entry
ABL1_CAEEL
> first['SEQ']; # get the sequence for the 1st entry
NNEWCEARLYSTRKNDASNQRRLGEIGWVPSNFIAPYNSLDKYTWYHGKI
..(557).. DVVPLAEKNVR
> second['FT'];
ACT_SITE 409 409
> last_three['DE']; # get the description tag
#for the last three entries
[PROTO-ONCOGENE TYROSINE-PROTEIN KINASE YRK (EC 2.7.1.112) (P60-YRK) (YES REL\
ATED KINASE).,
TYROSINE-PROTEIN KINASE ZAP-70 (EC 2.7.1.112) (70 KD ZETA-ASSOCIATED PROTEIN).,
TYROSINE-PROTEIN KINASE ZAP-70 (EC 2.7.1.112) (70 KD ZETA-ASSOCIATED PROTEIN).]
Notice that when an Entry structure has only a single posint parameter, as is the case with first and second
above, and we select for a specific tag, then it returns the
contents contained in this field as a name object.
When more than one entry is specified, as is the case with last_three, it returns a list of string objects. The
ith element of this list corresponds to the ith parameter of
Entry.8.1
Figure:
A
diagram showing the database structure DB. Here
DB is a protein database consisting of 78 sequences from
Swiss-Prot. The newline characters have been changed to
space symbols.
 |



Next: The Offset Structure
Up: Accessing a Darwin Sequence
Previous: Accessing a Darwin Sequence
Gaston Gonnet
1998-09-15
![[*]](cross_ref_motif.gif) gives a graphical view of how information is organized internally.
DB[string] points to the beginning of this name.
Recall that each entry from a sequence database in Darwin is wrapped
in the SGML tags <E>, </E>. To extract the entire
contents of an entry, we use the Entry structured type.
gives a graphical view of how information is organized internally.
DB[string] points to the beginning of this name.
Recall that each entry from a sequence database in Darwin is wrapped
in the SGML tags <E>, </E>. To extract the entire
contents of an entry, we use the Entry structured type. 